Center for Infection and Immunity
The Center for Infection and Immunity (CII) is one of the world's most advanced academic laboratories focused on pathogen discovery, infectious disease surveillance, and identifying the cause of chronic illnesses. It is a longstanding member of the WHO Global Outbreak Response Network (GOARN), originating collaborator with the Norwegian Institute of Public Health in the Autism Birth Cohort Study (ABC) , and is the home to the Global Alliance for Preventing Pandemics (GAPP) as well as the Center for Solutions for ME/CFS.
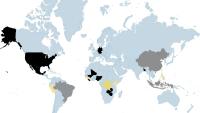
The Center's pioneering molecular methods in discovery have identified more than 2,000 viruses amongst other microbial threats with methods for accurate screening and diagnostics for these agents. The center has shepherded over a combined 60 postdoctoral fellows and graduate students, as well as hosted multiple sabbatical professors and trained more than 50 investigators from 30 countries in pathogen discovery and diagnostics throughout its history. The CII works at the forefront of outbreaks and is committed to the service of global public health, collaborating with scientists from nearly 50 different countries spanning six continents.
Our aims are to:
- Determine the roles of microbes in the development of acute infectious and chronic diseases
- Dissect the mechanisms by which the response to infection results in disease
- Find strategies to reduce the global burden of infectious diseases
Support the Center for Infection and Immunity
Our work would not be possible without the generous support of our partners